Synechococcus
 The marine cyanobacteria of the genus Synechococcus carry out a large portion of carbon fixation in the oceans, and their genomes are among the most abundant on the planet. Together with their relatives from the genus Prochlorococcus, these photosynthetic prokaryotic cells are the predominant members of the marine phytoplankton, which is responsible for about half of the world’s photosynthesis.
The marine cyanobacteria of the genus Synechococcus carry out a large portion of carbon fixation in the oceans, and their genomes are among the most abundant on the planet. Together with their relatives from the genus Prochlorococcus, these photosynthetic prokaryotic cells are the predominant members of the marine phytoplankton, which is responsible for about half of the world’s photosynthesis.
Therefore, the study of these organisms is of great biological and ecological interest. Although the genus Synechococcus is less abundant than Prochlorococcus on a global scale, it is more ubiquitous and more diverse. In tropical and equatorial zones, both genera co-inhabit the central zones of the oceans which are characterized by a lack of mineral elements (oligotrophy).
Synechococcus prefers well-lit, nutrient-rich waters and usually blooms in spring when the water column is mixed. The Synechococcus group is phylogenetically split into 3 main branches, called sub-clusters 5.1 to 5.3. The most abundant and diversified one is seemingly the sub-cluster 5.1 in which at least 9 clades have been defined using the 16S rDNA gene (Fuller et al., 2003) and 28 using the high resolution marker petB (Mazard et al., 2012 6). Clades I and IV generally co-occur at latitudes above 30°N/S and seem to be restricted to near coastal waters (Zwirglmaier et al., 2007; Zwirglmaier et al., 2008 12; Tai and Palenik, 2009; Mazard et al., 2011; Mella-Flores et al., 2012 8), whereas clade II seems to be abundant in warm, coastal or shelf areas of (sub)tropical waters (Zwirglmaier et al., 2008 12). The latitudinal distribution of clade III appears to be broader, but cells belonging to this ecotype seemingly prefer oligotrophic, offshore waters. All other clades are generally found at lower concentrations than clades I to IV and their distribution patterns are therefore less clearly defined (Zwirglmaier et al., 2008 12). Marine Synechococcus are divided into three major pigment types with PBS rods of type 1 containing only phycocyanin (PC), type 2 both PC and phycoerythrin I (PEI), and type 3 PC, PEI and PEII (Six et al., 2007 11). Type 3 can be further split into four subtypes (3a-d) based on the ratio of the phycourobilin (PUB) to phycoerythrobilin (PEB) chromophores bound to PEs. Pigment type 3d is able to vary its pigmentation in response to changes in ambient light color through a process called Type IV chromatic acclimation (Everroad et al., 2006, Six et al., 2007 11).
Useful links
Genome browser
Orthologous sequences (Cyanorak)
Order mutants
Genetic resources
Available data
Available individuals
- Public genomes
- All genomes
Toolbox
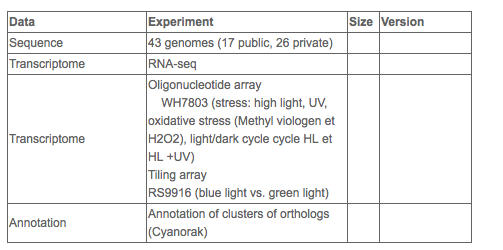
- 43 complete genome sequences
- Extensive transcriptome data (oligonucléotide-based microarray data, whole genome tiling array data)
- Mutant screen methodologies: flow cytometry, spectrofluorimetry, PAM fluorimetry
- Bioinformatic tools (Cyanorak Information system)
- ~350 strains in the Roscoff Culture Collection (http://www.sb-roscoff.fr/Phyto/RCC/) representative of the wide genetic diversity and pigmentation as well as from various oceanic regimes.
- Cryopreservation protocol
- Classical genetic methods (conjugation)
- Brahamsha, B. (1996). A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Applied and Environmental Microbiology 62, 1747-1751.
- McCarren, J., and Brahamsha, B. (2005). Transposon mutagenesis in a marine Synechococcus strain: isolation of swimming motility mutants. J Bacteriol 187, 4457-4462.
- Plasmids available :
- Replicative plasmid: pRL153 (RSF1010, RSF1010, IncQ, kanR)
- Integrative plasmid: pMUT100 (pBR322, pMB1, TetR et KmR ou Strep/SpecR)
- Conjugative plasmids: pRK24 (RP4, IncP, RK2, TetR AmpR KanR); pRK2013 (IncP, RK2/ColE1, kanR)
- Helper plasmid: pRL528 (colK, porte les gènes mob, CmR)
- Transposon plasmid: pRL27 (mini-Tn5 plasposon, KanR)
Culture:
Culture media used to cultivate marine Synechococcus are described in the Roscoff Culture Collection.
Contact:
Dr. Laurence Garczarek, Marine Phototrophic Prokaryotes (MaPP) team, UMR 7144 CNRS-UPMC, Station Biologique, Place Georges Teissier, CS 90074, 29688 Roscoff, France
External links:
Marine Synechococcus genomes were sequenced by:
- The Joint Genome Institute (JGI)
- The J. Craig Venter Institute (JCVI)
- The Genoscope
Other useful links:
- The Marine Prototrophic Prokaryotes (MaPP) team
- The Roscoff Oceanic Plankton group
- The Roscoff Culture Collection (RCC)
References:
- Blot, N., Mella-Flores, D., Six, C., Le Corguillé, G., Boutte, C., Peyrat, A., Monnier, A., Ratin, M., Gourvil, P., Campbell, D.A., and Garczarek, L. (2011). Light history influences the response of the marine cyanobacterium Synechococcus sp. WH7803 to oxidative stress. Plant Physiol 156, 1934-1954.
- Bretaudeau, A., Coste, F., Humily, F., Garczarek, L., Le Corguillé, G., Six, C., Ratin, M., Collin, O., Schluchter, W.M., and Partensky, F. (2013). CyanoLyase: a database of phycobilin lyase sequences, motifs and functions. Nucleic Acids Res 41, D396-401.
- Coelho, S. M., Simon, N., Ahmed, S., Cock, J. M. & Partensky, F. 2013. Ecological and evolutionary genomics of marine photosynthetic organisms. Molecular Ecology 22:867-907
- Dufresne, A., Ostrowski, M., Scanlan, D.J., Garczarek, L., Mazard, S., Palenik, B.P., Paulsen, I.T., De Marsac, N.T., Wincker, P., Dossat, C., Ferriera, S., Johnson, J., Post, A.F., Hess, W.R., and Partensky, F. (2008). Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol 9, R90.
- Garczarek, L., Dufresne, A., Blot, N., Cockshutt, A.M., Peyrat, A., Campbell, D.A., Joubin, L., and Six, C. (2008). Function and evolution of the psbA gene family in marine Synechococcus: Synechococcus sp. WH7803 as a case study. Isme J 2, 937-953.
- Mazard, S., Ostrowski, M., Partensky, F., and Scanlan, D.J. (2012). Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environ Microbiol 14, 372-386.
- Mella-Flores, D., Mazard, S., Humily, F., Partensky, F., Mahé, F., Bariat, L., Courties, C., Marie, D., Ras, J., Mauriac, R., Jeanthon, C., Bendif, E.M., Ostrowski, M., Scanlan, D.J., and Garczarek, L. (2011). Is the distribution of Prochlorococcus and Synechococcus ecotypes in the Mediterranean Sea affected by global warming? Biogeosciences 8, 2785–2804
- Mella-Flores, D., Six, C., Ratin, M., Partensky, F., Boutte, C., Le Corguillé, G., Marie, D., Blot, N., Gourvil, P., Kolowrat, C., and Garczarek, L. (2012). Prochlorococcus and Synechococcus have Evolved Different Adaptive Mechanisms to Cope with Light and UV Stress. Front Microbiol 3, 285.
- Scanlan, D.J., Ostrowski, M., Mazard, S., Dufresne, A., Garczarek, L., Hess, W.R., Post, A.F., Hagemann, M., Paulsen, I., and Partensky, F. (2009). Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73, 249-299.
- Shukla, A., Biswas, A., Blot, N., Partensky, F., Karty, J.A., Hammad, L.A., Garczarek, L., Gutu, A., Schluchter, W.M., and Kehoe, D.M. (2012). Phycoerythrin-specific bilin lyase-isomerase controls blue-green chromatic acclimation in marine Synechococcus. Proc Natl Acad Sci U S A 109, 20136-20141.
- Six, C., Thomas, J.C., Garczarek, L., Ostrowski, M., Dufresne, A., Blot, N., Scanlan, D.J., and Partensky, F. (2007). Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study. Genome Biol 8, R259.
- Zwirglmaier, K., Jardillier, L., Ostrowski, M., Mazard, S., Garczarek, L., Vaulot, D., Not, F., Massana, R., Ulloa, O., and Scanlan, D.J. (2008). Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol 10, 147-161.





